
| Size | Price | Stock | Qty |
|---|---|---|---|
| 5mg |
|
||
| 10mg |
|
||
| 25mg |
|
||
| 50mg |
|
||
| 100mg |
|
||
| 250mg |
|
||
| Other Sizes |
Purity: =99.05%
VE-821 is a novel potent and highly selective ATP competitive protein kinase inhibitor of ATR (ataxia telangiectasia mutated and Rad3 related) with Ki and IC50 of 13 nM and 26 nM in cell-free assays, it shows inhibition of H2AX phosphorylation, and had minimal activity against PIKKs ATM, DNA-PK, mTOR and PI3Kγ. VE-821 confirmed that ATR signaling was inhibited by preventing Chk1 from being phosphorylated in response to radiation and gemcitabine. Under both normoxic and hypoxic conditions, VE-821 consistently increased the susceptibility of PSN-1, MiaPaCa-2, and primary PancM pancreatic cancer cells to radiation and gemcitabine.
| Targets |
ATR ( Ki = 13 nM ); ATM ( Ki = 16 μM ); DNA-PK ( Ki = 2.2 μM ); PI3Kγ ( Ki = 3.9 μM )
|
||
|---|---|---|---|
| ln Vitro |
|
||
| ln Vivo |
|
||
| Enzyme Assay |
Radiometric-phosphate incorporation assay is used to determine a compound's ability (e.g., VE-821) to inhibit ATR, ATM, or DNAPK kinase activity. After the proper buffer, kinase, and target peptide are combined, a stock solution is created. To achieve a final DMSO concentration of 7%, the compound of interest is added to this at different DMSO concentrations. When the proper [g-33P]ATP solution is added, the assay is started and incubated at 25°C. The assays are terminated by adding phosphoric acid and ATP to a final concentration of 100 mM and 0.66 μM, respectively, following the desired time course. Peptides are prepared on a phosphocellulose membrane, captured, and then six times washed with 200 μL of 100 mM phosphoric acid. Next, 100 μL of scintillation cocktail is added, and the sample is counted using a 1450 Microbeta Liquid Scintillation Counter. GraphPad Prism software is used to analyze dose-response data[2].
|
||
| Cell Assay |
Plated in 96-well plates, MiaPaCa-2, PSN-1, and Panc1 cells (5×104) are treated with increasing concentrations of VE-821 after 4 hours, and 1 hour before they are exposed to a single 4 Gy dose of radiation. After the medium is changed 72 hours after the radiation, the Alamar Blue assay is used to determine viability. After allowing the cells to multiply, the viability of the cells is examined once more on day 10 for each of the various treatment scenarios. Normalization of cell viability and survival fraction to the untreated (control) group is achieved [3].
Cell lines were purchased from ATCC and maintained according to the distributor's instructions. Cell assays were performed using exponentially growing cultures. For H2AX phosphorylation analysis using immunofluorescence (IF) microscopy, cells were fixed in 4% formaldehyde, permeabilized with 0.5% Triton X-100, and stained with mouse H2AX pS139 antibody, AlexaFluor 488 goat antimouse antibody, and Hoechst. The cells were then analyzed using the BD Pathway 855 bioimager and BD Attovision software. The cell density was analyzed using the CellTiter 96 AQueous Cell Proliferation (MTS) assay. Cells were plated in 96-well plates and allowed to adhere overnight. The following day, compounds were added at the indicated concentrations in a final volume of 200 μL, and the cells were then incubated for 96 h. MTS reagent (40 μL) was then added, and 1 h later, absorbance at 490 nm was measured using a SpectraMax Plus 384 plate reader. Synergy and antagonism were assessed using Macsynergy software.[2] |
||
| Animal Protocol |
|
||
| References |
|
||
| Additional Infomation |
3-amino-6-(4-methylsulfonylphenyl)-N-phenyl-2-pyrazinecarboxamide is an aromatic amide.
DNA-damaging agents are among the most frequently used anticancer drugs. However, they provide only modest benefit in most cancers. This may be attributed to a genome maintenance network, the DNA damage response (DDR), that recognizes and repairs damaged DNA. ATR is a major regulator of the DDR and an attractive anticancer target. Herein, we describe the discovery of a series of aminopyrazines with potent and selective ATR inhibition. Compound 45 inhibits ATR with a K(i) of 6 nM, shows >600-fold selectivity over related kinases ATM or DNA-PK, and blocks ATR signaling in cells with an IC(50) of 0.42 μM. Using this compound, we show that ATR inhibition markedly enhances death induced by DNA-damaging agents in certain cancers but not normal cells. This differential response between cancer and normal cells highlights the great potential for ATR inhibition as a novel mechanism to dramatically increase the efficacy of many established drugs and ionizing radiation.[2] DNA damaging agents such as radiotherapy and gemcitabine are frequently used for the treatment of pancreatic cancer. However, these treatments typically provide only modest benefit. Improving the low survival rate for pancreatic cancer patients therefore remains a major challenge in oncology. Inhibition of the key DNA damage response kinase ATR has been suggested as an attractive approach for sensitization of tumor cells to DNA damaging agents, but specific ATR inhibitors have remained elusive. Here we investigated the sensitization potential of the first highly selective and potent ATR inhibitor, VE-821, in vitro. VE-821 inhibited radiation- and gemcitabine-induced phosphorylation of Chk1, confirming inhibition of ATR signaling. Consistently, VE-821 significantly enhanced the sensitivity of PSN-1, MiaPaCa-2 and primary PancM pancreatic cancer cells to radiation and gemcitabine under both normoxic and hypoxic conditions. ATR inhibition by VE-821 led to inhibition of radiation-induced G 2/M arrest in cancer cells. Reduced cancer cell radiosurvival following treatment with VE-821 was also accompanied by increased DNA damage and inhibition of homologous recombination repair, as evidenced by persistence of γH2AX and 53BP1 foci and inhibition of Rad51 foci, respectively. These findings support ATR inhibition as a novel approach to improve the efficacy and therapeutic index of standard cancer treatments across a large proportion of pancreatic cancer patients.[3] Inhibiting the bromodomain and extra-terminal (BET) domain family of epigenetic reader proteins has been shown to have potent anti-tumoral activity, which is commonly attributed to suppression of transcription. In this study, we show that two structurally distinct BET inhibitors (BETi) interfere with replication and cell cycle progression of murine Myc-induced lymphoma cells at sub-lethal concentrations when the transcriptome remains largely unaltered. This inhibition of replication coincides with a DNA-damage response and enhanced sensitivity to inhibitors of the upstream replication stress sensor ATR in vitro and in mouse models of B-cell lymphoma. Mechanistically, ATR and BETi combination therapy cause robust transcriptional changes of genes involved in cell death, senescence-associated secretory pathway, NFkB signaling and ER stress. Our data reveal that BETi can potentiate the cell stress and death caused by ATR inhibitors. This suggests that ATRi can be used in combination therapies of lymphomas without the use of genotoxic drugs.[4] |
| Molecular Formula |
C18H16N4O3S
|
|
|---|---|---|
| Molecular Weight |
368.41
|
|
| Exact Mass |
368.094
|
|
| Elemental Analysis |
C, 58.68; H, 4.38; N, 15.21; O, 13.03; S, 8.70
|
|
| CAS # |
1232410-49-9
|
|
| Related CAS # |
|
|
| PubChem CID |
51000408
|
|
| Appearance |
White to beige solid powder
|
|
| Density |
1.4±0.1 g/cm3
|
|
| Boiling Point |
568.4±50.0 °C at 760 mmHg
|
|
| Flash Point |
297.6±30.1 °C
|
|
| Vapour Pressure |
0.0±1.6 mmHg at 25°C
|
|
| Index of Refraction |
1.658
|
|
| LogP |
2.93
|
|
| Hydrogen Bond Donor Count |
2
|
|
| Hydrogen Bond Acceptor Count |
6
|
|
| Rotatable Bond Count |
4
|
|
| Heavy Atom Count |
26
|
|
| Complexity |
578
|
|
| Defined Atom Stereocenter Count |
0
|
|
| SMILES |
S(C([H])([H])[H])(C1C([H])=C([H])C(C2=C([H])N=C(C(C(N([H])C3C([H])=C([H])C([H])=C([H])C=3[H])=O)=N2)N([H])[H])=C([H])C=1[H])(=O)=O
|
|
| InChi Key |
DUIHHZKTCSNTGM-UHFFFAOYSA-N
|
|
| InChi Code |
InChI=1S/C18H16N4O3S/c1-26(24,25)14-9-7-12(8-10-14)15-11-20-17(19)16(22-15)18(23)21-13-5-3-2-4-6-13/h2-11H,1H3,(H2,19,20)(H,21,23)
|
|
| Chemical Name |
3-amino-6-(4-methylsulfonylphenyl)-N-phenylpyrazine-2-carboxamide
|
|
| Synonyms |
VE-821; VE 821; 1232410-49-9; 3-Amino-6-(4-(methylsulfonyl)phenyl)-N-phenylpyrazine-2-carboxamide; VE 821; VE821; 3-Amino-6-[4-(methylsulfonyl)phenyl]-N-phenyl-2-pyrazinecarboxamide; BF884TQ935; 3-amino-6-(4-methylsulfonylphenyl)-N-phenylpyrazine-2-carboxamide; VE821
|
|
| HS Tariff Code |
2934.99.9001
|
|
| Storage |
Powder -20°C 3 years 4°C 2 years In solvent -80°C 6 months -20°C 1 month |
|
| Shipping Condition |
Room temperature (This product is stable at ambient temperature for a few days during ordinary shipping and time spent in Customs)
|
| Solubility (In Vitro) |
|
|||
|---|---|---|---|---|
| Solubility (In Vivo) |
|
| Preparing Stock Solutions | 1 mg | 5 mg | 10 mg | |
| 1 mM | 2.7144 mL | 13.5718 mL | 27.1437 mL | |
| 5 mM | 0.5429 mL | 2.7144 mL | 5.4287 mL | |
| 10 mM | 0.2714 mL | 1.3572 mL | 2.7144 mL |
*Note: Please select an appropriate solvent for the preparation of stock solution based on your experiment needs. For most products, DMSO can be used for preparing stock solutions (e.g. 5 mM, 10 mM, or 20 mM concentration); some products with high aqueous solubility may be dissolved in water directly. Solubility information is available at the above Solubility Data section. Once the stock solution is prepared, aliquot it to routine usage volumes and store at -20°C or -80°C. Avoid repeated freeze and thaw cycles.
Calculation results
Working concentration: mg/mL;
Method for preparing DMSO stock solution: mg drug pre-dissolved in μL DMSO (stock solution concentration mg/mL). Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug.
Method for preparing in vivo formulation::Take μL DMSO stock solution, next add μL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O,mix and clarify.
(1) Please be sure that the solution is clear before the addition of next solvent. Dissolution methods like vortex, ultrasound or warming and heat may be used to aid dissolving.
(2) Be sure to add the solvent(s) in order.
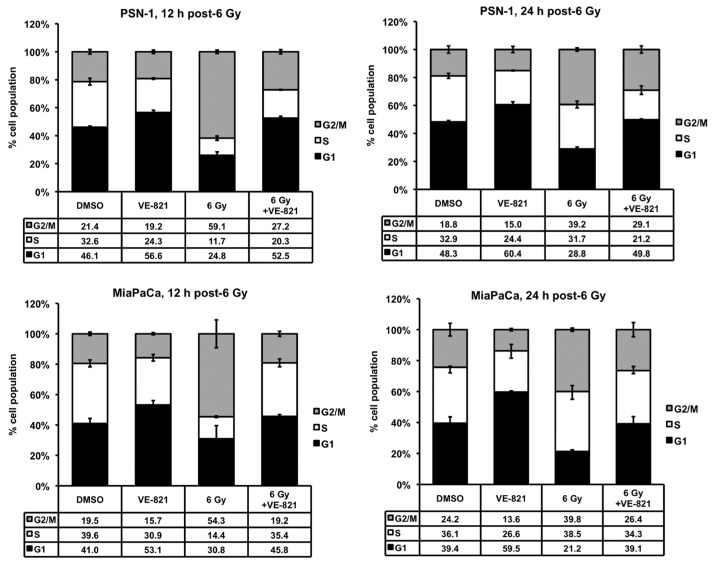 VE-821 perturbs the irradiation-induced cell cycle checkpoint in pancreatic cancer cells.Cancer Biol Ther.2012 Sep;13(11):1072-81. |
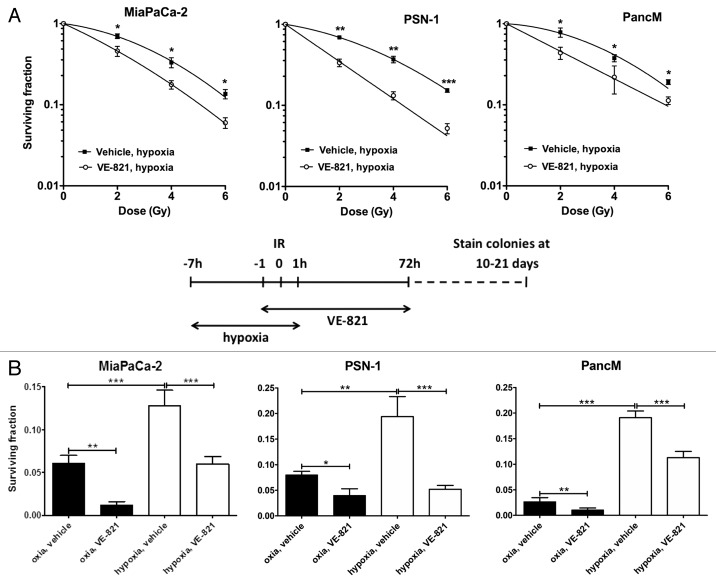 VE-821 radiosensitizes pancreatic tumor cells under hypoxic conditions.Cancer Biol Ther.2012 Sep;13(11):1072-81. |
VE-821 radiosensitizes pancreatic tumor cells.Cancer Biol Ther.2012 Sep;13(11):1072-81. |
|---|
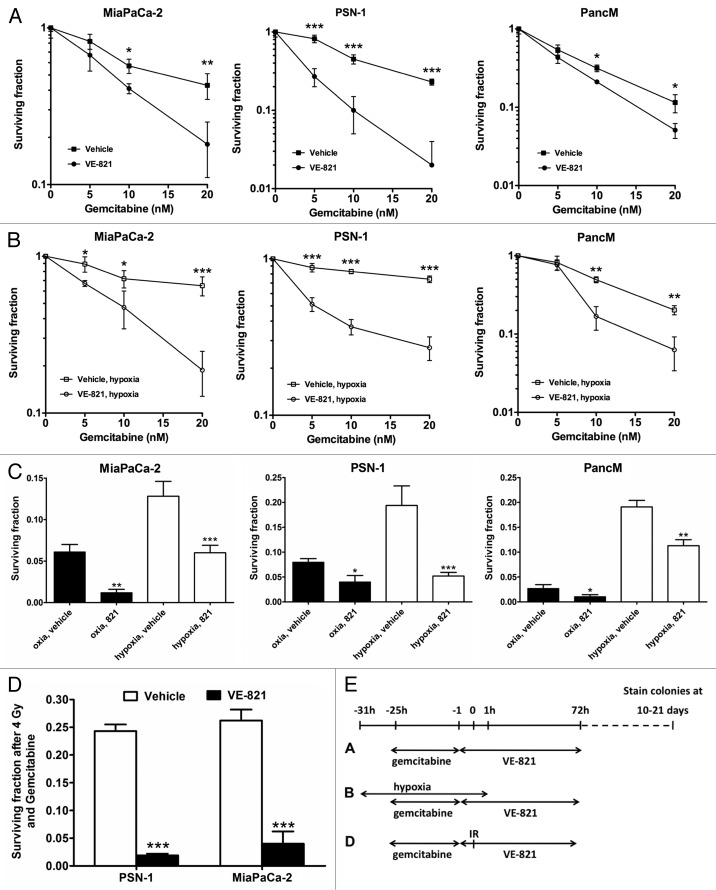 VE-821 sensitizes pancreatic cancer cells to gemcitabine treatment.Cancer Biol Ther.2012 Sep;13(11):1072-81. |
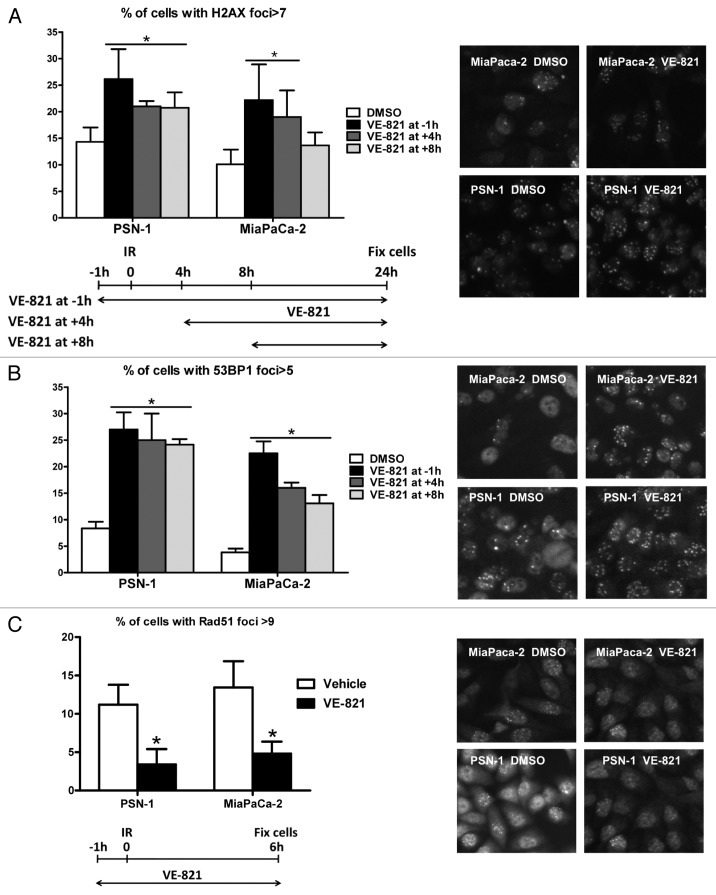 VE-821 increases 53BP1 and γH2AX foci number and reduces Rad51 foci formation.Cancer Biol Ther.2012 Sep;13(11):1072-81. |